Introduction of m6A
N6-methyladenosine (m6A) is the most frequently occurring
modification among more than 160 modifications.
It exists in eukaryotes including yeast, insects and mammals.
m6A is a DNA and RNA base modification at the 6th nitrogen position.
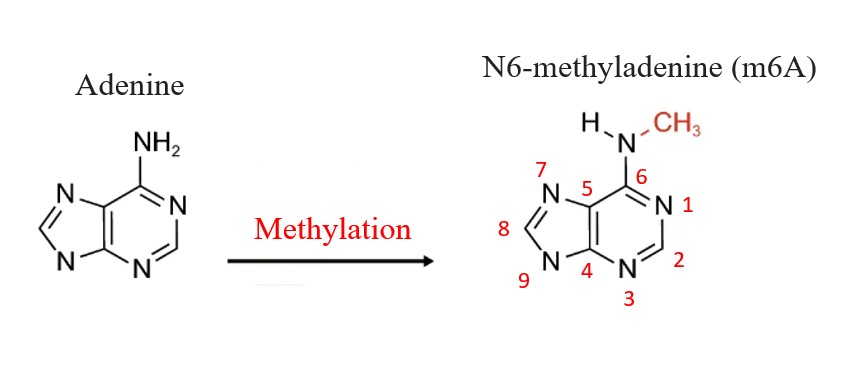 Recently, it
has been resurfaced as a potential reversible epigenetic mark in eukaryotes.
m6A can affect multiple biological processes including pairing between
the bases, positioning and stability of the nucleosome, replication and mismatch repair, signaling to
host-pathogen interaction, transcript synthesis, X-chromosome inactivation, gene imprinting, tissue
specific gene expression, virus resistance and epigenetic memory. Therefore, it is quite necessary to understand the mechanism of
m6A .
Recently, it
has been resurfaced as a potential reversible epigenetic mark in eukaryotes.
m6A can affect multiple biological processes including pairing between
the bases, positioning and stability of the nucleosome, replication and mismatch repair, signaling to
host-pathogen interaction, transcript synthesis, X-chromosome inactivation, gene imprinting, tissue
specific gene expression, virus resistance and epigenetic memory. Therefore, it is quite necessary to understand the mechanism of
m6A .
Various experiments have been conducted to identify m6A sites. The experiments are costly and time-consuming, as a result, lot of works utilize deep learning method to predict m6A sites on a large scale. Here we construct a database which provide simple access to previous research for users.
We roughly split previous works into two parts.
-
Algorithm, every work proposed their own model to predict whether the input is m6A or not. We provide the outline of each work, if paper does not provide their own code, we will reconstruct their model depend on their description.
-
Dataset, most of the datasets were made by experimental statistic, however, each dataset is screened out the data by different method. We give the detail of each dataset including length, positive data number, negative data number, etc.
N6-methyladenosine (m6A) is the most frequently occurring
modification among more than 160 modifications.
It exists in eukaryotes including yeast, insects and mammals.
m6A is a DNA and RNA base modification at the 6th nitrogen position.

Statistics of m6A
(I) Statistics of the 26 collected deep learning based m6A prediction studies.
Classify the studies by year
Classify the studies by web and code availability
(II) Statistics of the 52 collected m6A-related datasets.